Appendix D: Ancient DNA
Robyn Humphreys, MSc., University of Cape Town
This appendix is a revision of the “Chapter 11 Special Topics: Ancient DNA” by Robyn Humphreys. In Explorations: An Open Invitation to Biological Anthropology, first edition, edited by Beth Shook, Katie Nelson, Kelsie Aguilera, and Lara Braff, which is licensed under CC BY-NC 4.0.
Learning Objectives
- Describe the challenges in recovering and sequencing ancient DNA.
- Explain how the Denisovans were discovered and what we have learned about them based on their aDNA.
- Describe the relationships between Neanderthals, Denisovans, and modern humans based on aDNA evidence.
- Explain how DNA can provide insights into the population structure of hominin groups of the past.
Ancient DNA (aDNA) has provided us with new insights into our evolutionary history that cannot be garnered from the fossil record alone. For example, it has assisted with the discovery of the Denisovans, for whom little fossil evidence is available. It has helped us better understand, and make inferences about, the evolution of and relationships among Neanderthals, Denisovans, and modern humans. It has also helped to answer some very important questions about what happened when modern humans migrated out of Africa and encountered these European/Asian hominins, as we will discuss in this appendix.
Sequencing Ancient Genomes
The first successful sequencing of aDNA from an archaic hominin took place in 1997 with the sequencing of mitochondrial DNA (mtDNA) from a Neanderthal-type specimen from Feldhofer Cave. mtDNA is ideal for aDNA studies because it is more abundant than nuclear DNA in our cells. This mitochondrial sequence provided evidence that Neanderthals belonged in a clade separate from modern humans and that their mtDNA was four times more different from modern humans than modern human mtDNA was from each other (Krings et al. 1997).
Sequencing of nuclear DNA would not occur until more than ten years later. The first nuclear genomic sequence representing Neanderthals was produced by sequencing three individuals and using their sequences to create a composite draft Neanderthal genome (Green et al. 2010). The first high-coverage sequence of a single Neanderthal was that of a female Neanderthal who lived in Siberia, followed by another high-coverage sequence from a female Neanderthal whose remains were found in the Vidja cave in Croatia (Prüfer et al. 2014). High-coverage sequences are produced when the genome has been sequenced multiple times, which ensures that the sequences are a true reflection of the genomic sequence and not due to errors that occur during the process of sequencing.
Collecting and Sequencing aDNA
While aDNA can be collected from many different sources (e.g., soft tissue, hair, paleo feces, soils, and sediments), when studying ancient hominins it is most often collected from bone and teeth. Because extraction of aDNA requires destruction of part of the tissue, and the morphology of the skeletal element might be informative, care needs to be taken when deciding what is sampled. Multiple samples are often taken to allow repeat sequencing and demonstrate reproducibility of results. All samples must be minimally handled to avoid contamination.
Endogenous aDNA, or DNA that was present in the tissue before the body decomposed, are usually in fragments 100 to 300 base pairs (bp) long due to degradation, and thus difficult to study. Sometimes DNA from other sources, known as exogenous DNA, are also found in samples. Some examples include DNA from microbes or modern human contamination (Figure D.1).
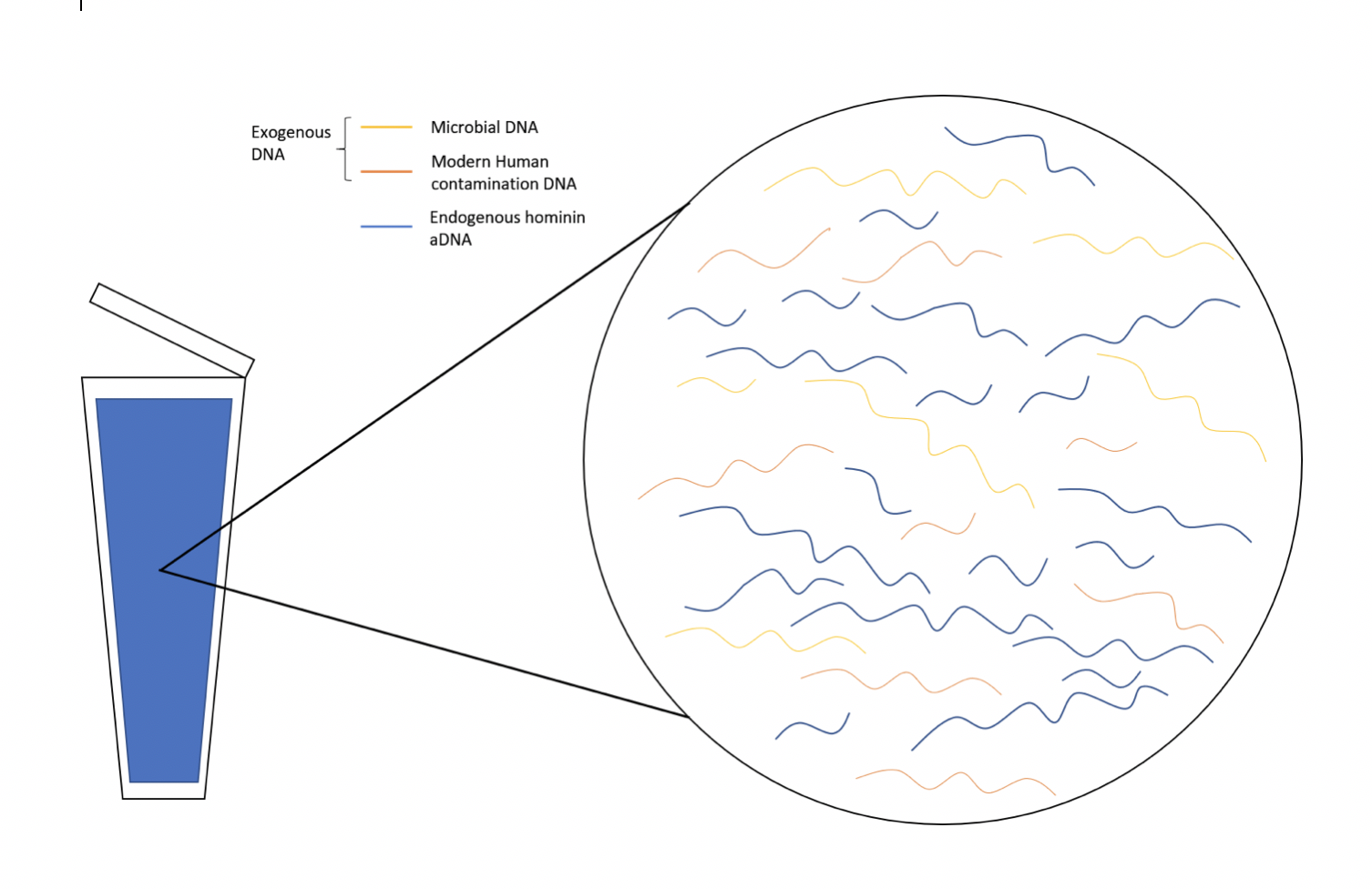
There are also modifications that occur to aDNA due to chemical reactions. For example, deamination results in Cytosine (C) to Thymine (T) conversions, which occur mostly at the 5’ end (5 prime end) of the DNA fragment. This in turn results in Guanine (G) to Adenine (A) substitutions on the 3’ end (3 prime end) of the DNA fragment. These sequence changes in aDNA might not reflect the original hominin sequence, yet these changes can be helpful when differentiating between aDNA and modern human DNA contamination. The environment plays a significant role, as DNA preserves well in cold conditions such as permafrost. aDNA has also been recovered from material found in drier environments under special conditions. Factors such as water percolation, salinity, pH, and microbial growth all affect the preservation of aDNA.
The bone that best preserves DNA after death is the petrous portion of the temporal bone. This forms part of the skull and protects the inner ear. Due to its high density, the petrous portion preserves DNA very well. Thus, it is possible to get DNA from older and less well-preserved individuals when using the petrous portion. Compared to other bones, the petrous portion not only preserves DNA better but also allows for the extraction of more DNA. The petrous portion can yield up to 100 times more DNA than other bones (Pinhasi et al. 2015)
Initially the short fragments and degraded nature of aDNA posed a big problem with the usual polymerase chain reaction (PCR) procedures used to sequence DNA. But, the advent of high-throughput sequencing has revolutionized sequencing the genomes of ancient hominins. High-throughput sequencing allows for the parallel sequencing of many fragments of DNA in one reaction, without prior knowledge of the target sequence. In this way, the maximum amount of available aDNA can be sequenced. Because the high-throughput sequencing method does not discriminate between endogenous aDNA and exogenous DNA, it is important to either ensure that there is as little contamination as possible and/or use methods that allow for differentiation of the target aDNA.
The Discovery of Denisovans
Denisovans, named after the Siberian cave in which they were discovered, are a distinct group of hominins that were identified through aDNA. Analysis of the ancient mtDNA from teeth and bone fragments revealed they had haplotypes outside the range of variation of modern humans and Neanderthals. The phalanx bone from which the DNA of the Denisovan was recovered did not have traits that indicated that it was another species. A haplotype is a set of genetic variants located on a single stretch of the genome. Shared or similar haplotypes can be used to identify ancestral relationships and to differentiate groups. Dubbed lineage X, the mtDNA sequence from these fossils suggested that Denisovans diverged from modern humans and Neanderthals.
Subsequent high-coverage sequence of a Denisovan (Denisovan 3) nuclear genome showed that Denisovans are a sister group to Neanderthals and thus more closely related to them than indicated by the mtDNA data (Brown et al. 2016). Because mtDNA and nuclear DNA have different patterns of inheritance, they can paint different pictures about the relationships between two groups. The Denisovans are now thought to have a mtDNA sequence derived from an ancient hominin group that hybridized with Denisovans and introduced the mtDNA sequence.
Sequences from three other Denisovans (Denisovan 2, 4, and 8) that provide insight into how old the specimens are, along with the usual dating methods (such as radio carbon dating and uranium dating), show that Denisovans occupied the Denisovan cave from around 195 kya to 52 kya to 76 kya. DNA can assist with dating because, compared to older sequences, younger sequences will have accumulated more mutations from the putative common ancestral sequence. Thus, it is possible to conclude from sequence data, that Denisovan 2 is 54.2 kya to 99.4 kya older than Denisovan 3, and 20.6 kya to 37.7 kya older than Denisovan 8. Molecular data indicates that Neanderthals and Denisovans separated between 381 kya and 473 kya and that the branch leading to Denisovans and modern humans diverged around 800 kya. Denisovans are also more closely related to another set of fossils found in the cave Sima de los Huesos dated to 430 kya. Thus, the split between Neanderthals and Denisovans must have occurred before 430 kya (Meyer et al. 2016).
What Can We Learn About Population Structure of the Neanderthals and Denisovans from aDNA?
Ancient DNA has helped us understand the demographics of Neanderthals and Denisovans and make inferences about population size and history. Three lines of evidence suggest that these groups had small populations toward the end of their existence.
The first line of evidence uses coalescent methods. This process is used to determine which population dynamics in the past are most likely to give rise to the genetic sequences we have, and it allows us to understand population changes of the past, including recombination, population subdivision, and variable population size.
The second indicator that Neanderthals and Denisovans had smaller population sizes is that these groups carried many deleterious (or harmful) genomic variants. Genomic variants are considered deleterious when the change in genomic sequence alters the amino acid sequence of a protein and affects the function of the protein; such variants are known as nonsynonymous mutations. By contrast, synonymous mutations that occur in protein-coding regions of the genome do not change the amino acid sequence nor affect the proteins produced. Denisovans and Neanderthals have a higher ratio of nonsynonymous to synonymous mutations when compared to contemporary modern human populations. This is indicative of a small population size, because if the population were larger, natural selection would have acted on these deleterious variants and weeded them out.
A third indicator of small population size is that the Neanderthals sequenced thus far have low levels of heterozygosity. Heterozygosity is measured by looking at how often two different alleles are found within a certain stretch of DNA. When you find many regions on the genome with different alleles, there is a high level of heterozygosity. When you find very few positions where there are two different alleles, this indicates a low level of heterozygosity. Both Neanderthals and Denisovans appear to have low levels of heterozygosity, indicating smaller population sizes. Ancient Neanderthal genomes also revealed that there were consanguineous relations (mating relationships between two closely related Neanderthals). This was determined by looking at the stretches of homozygosity in a female neanderthal’s genome that were longer than expected and could not be explained by small population size alone.
Sequencing Archaic Genomes to Understand Modern Humans
Not only did the sequencing of archaic genomes allow us to learn more about Neanderthals and Denisovans, it gave us important insights into our own evolution. Previously the human genome was only compared to our closest living relatives, the great apes, which helped us identify unique derived genomic changes that occurred since humans split from our last common ancestor with chimpanzees. Neanderthal and Denisovan genomes provide another set of comparative samples that might help us identify changes unique to modern humans that occurred after our split from the last common ancestor with Neanderthals/Denisovans. These changes may help account for our success as a species.
Hybridization Between Hominin Groups
aDNA also provides insight into interactions between modern humans migrating out of Africa and other hominins that evolved in Europe and Asia. One of the hypotheses tested was this: if hybridization between modern humans and Neanderthals occurred, Neanderthals would have shared more genomic variants with some modern human populations than with others. When this was tested, the data showed that Neanderthals shared more genomic variants with Europeans and Asians than with African individuals (Sankararaman 2016). This difference in relatedness was significant, indicating that there had been hybridization between Neanderthals and modern humans.
From the genetic data, we know that Europeans have a smaller proportion of Neanderthal-derived genes than East Asians do (Prüfer et al. 2017). Thus, there was more admixture into ancestral East Asian populations than into ancestral European populations. Oceanians (Melanesians, Australian aborigines, and other Southeast Asian islanders) have a higher proportion of their DNA derived from Denisovans and longer stretches of Denisovan DNA. DNA in chromosomes get exchanged and experience genetic recombination, whereby introgressed regions (inherited from different species or taxon) are broken down into smaller segments each generation. Thus, longer stretches of introgressed DNA indicate that hybridization occurred more recently. Genetic analysis shows that the admixture event between the Denisovan and human ancestors of these populations is more recent than the admixture events between Neanderthals and modern humans.
To determine whether shared sequences are a result of introgression or more ancient substructure, researchers use divergence time: a measure of how long two sequences have been changing independently. The longer the two sequences have been changing independently, the more differences they will accumulate, which will result in a longer divergence time. By measuring the divergence time between the introgressed regions in modern human genomes and the Neanderthal sequences, researchers can calculate that the shared sequences are recent as well as date to when the two taxa made secondary contact. This is well after the initial population split between modern humans and Neanderthals. There has been gene flow from Neanderthals and Denisovans into modern human populations, between Neanderthals and Denisovans, and from modern humans into Neanderthals.
There is variation in how much of the Neanderthal genome is represented in the modern human population. Individuals outside of Africa usually have 1% to 4% of their genome derived from Neanderthals. Approximately 30% of the Neanderthal genome is represented in modern human genomes, altogether.
Introgressed genes have signatures that allow us to identify them and differentiate them from parts of the genome that are not introgressed. This can be identified in at least three ways. First, in this case, if the sequence is more similar to the Neanderthal sequence (i.e.,fewer sequence differences from the Neanderthal than the African modern human), it is likely that it is derived from a Neanderthal. Second, what is the divergence time between the allele and the same allele in a Neanderthal? If it is shorter than the divergence time between humans and Neanderthal, then the gene is most likely introgressed. An example of this can be seen in Figure D.2. Third, consider whether the allele that meets the first two criteria and is identified as possibly being introgressed can be found at higher frequencies in populations outside of Africa.

Examining the genomes of modern humans, we can see that there are regions of the genome with no Neanderthal and Denisovan genomic variants. These are known as Neanderthal or Denisovan introgression deserts. There are also overlaps between regions in the human genome that are Neanderthal and Denisovan deserts, which might indicate genomic incompatibilities between modern humans and these groups, resulting in those genes being selected against in the modern human genome. We can also infer that hybridization may itself have been a barrier to gene flow because there is a significant reduction in introgression on the X chromosome and around genes that are disproportionately expressed in the testes compared to other tissue groups. This could indicate that hybridization between modern humans and Neanderthals may have resulted in male hybrid infertility.
Because of the climate in Africa, it has been difficult or impossible to extract aDNA from African fossil remains. However, analysis of genomes of modern African populations indicate that there was admixture between modern humans and other hominins within Africa (see Figure D.2).
Confirmed Fossil Hybrids
Another line of evidence concerns hybrids. A first-generation hybrid is called an F1 hybrid; it is the direct offspring of two lineages that have been evolving independently over an extended period. A second-generation hybrid (F2) would be the offspring of two F1 hybrids. A backcrossed individual is the result of an F1 or F2 hybrid mating with an individual from one of the parental populations. An example of a backcross would be when a Neanderthal-human hybrid produces offspring with a human; their offspring would be considered a first-generation backcrossed hybrid (B1). Sequencing of aDNA from fossil material has confirmed that hybridization between different hominins has occurred, supporting the introgression data from recent populations.
The sequencing of Oase 1, a suspected hybrid based on skeletal morphology, showed that it had a Neanderthal ancestor as recently as six to eight generations back. He would thus be considered a backcrossed individual. The recent sequencing of a 13-year-old Denisovan female showed that she was the F1 hybrid offspring of a Neanderthal mother (from whom she inherited Neanderthal mtDNA) and a Denisovan father. While these are only two examples of individuals who are confirmed hybrids, many other remains show some indication of gene flow between hominins.
The Future of Genetic Studies
We are continuing to learn how introgressed genes affect modern humans. Combining phenotypic and genetic information, Neanderthal-derived genes have been associated with diverse traits, ranging from thes skin’s sun sensitivity to excessive blood clotting by certain individuals. Interesting research has also shown that introgressed alleles might produce different gene expression profiles when compared to non-introgressed alleles. However, there is a lot of research still to be done to fully understand the effects of introgression on modern populations and how it might have assisted modern humans who migrated out of Africa.
Review Questions
- What are three reasons that ancient DNA is so difficult to study?
- What are introgressed regions of DNA? What insights do studying introgression provide about early hominins?
- Diagram our current understanding of Denisovan, Neanderthal, and modern human lineages based on ancient DNA.
- How can ancient DNA help us understand Neanderthal demographics?
Key Terms
5 prime end: A nucleic acid strand that terminates at the chemical group attached to the fifth carbon in the sugar-ring.
3 prime end: A nucleic acid strand that terminates at the hydroxyl (-OH) chemical group attached to the third carbon in the sugar-ring.
Allele: Each of two or more alternative forms of a gene that arise by mutation and are found at the same place on a chromosome.
Coalescent methods: These are models which allow for inference of how genetic variants sampled from a population may have originated from a common ancestor
Deamination: The chemical process that results in the conversion of Cytosine to uracil, which results in Cytosine to Thymine conversions during sequencing.
Divergence time: A measure of how long two genomic sequences have been changing independently.
Endogenous aDNA: A form of ancient DNA in which DNA originates from the specimen being examined.
Exogenous DNA: DNA that originates from sources outside of the specimen you are trying to sequence.
Genetic recombination: This is the process of exchange of DNA between two strands to produce new sequence arrangements.
Haplotype: A set of genetic variants located on a single stretch of the genome. This unique combination of variants on a stretch of the genome can be used to differentiate groups that will have different combinations of variants.
Heterozygosity: A measure of how many genes within a diploid genome are made up of more than one variant for a gene.
High-coverage sequences: These are genomic sequences which have been sequenced multiple times to ensure that the sequence produced is a true reflection of the genomic sequence, and reduce the likelihood that the sequence has sequencing errors as a result of the sequencing process.
High–throughput sequencing: DNA sequencing technologies developed in the early 21st century that are capable of sequencing many DNA molecules at a time.
Homozygosity: A measure of how many genes within a diploid genome are made up of more than the same variant for a gene.
Hybridization: Mating between two genetically differentiated groups (or species).
Introgressed genes: This is the movement of genes from one species to the gene pool of another species through hybridization between the species and backcross into the parental population by hybrid offspring.
Nonsynonymous mutations: These are changes that occur in the protein-coding region of the genome and result in a change in amino acid sequence of the protein being produced.
Synonymous mutations: Mutations that occur in the protein-coding region of the genome but do not result in a change in amino acid sequence of the protein being produced.
About the Author
Robyn Humphreys, MSc.
University of the Western Cape, rhumphreys@uwc.ac.za
Robyn Humphreys is a biological anthropologist based in the archaeology department at the University of Cape Town. Her MSc focused on the role of hybridization in human evolution. She is now pursuing her Ph.D., which will involve looking at the relationship between archaeologists and communities in relation to research on human remains from historical sites in Cape Town.
For Further Exploration
Fu, Qiaomei, Mateja Hajdinjak, Oana Teodora Moldovan, Silviu Constantin, Swapan Mallick, Pontus Skoglund, Nick Patterson, et al. 2015. “An Early Modern Human from Romania with a Recent Neanderthal Ancestor.” Nature 524 (7564): 216.
Pääbo, Svante. 2011. “DNA Clues to Our Inner Neanderthal.,” TED Talk by Svante Pääbo, August 2011. Last accessed May 7, 2023. https://www.ted.com/talks/svante_paeaebo_dna_clues_to_our_inner_neanderthal?language=en.
References
Beyin, Amanuel. 2011. “Upper Pleistocene Human Dispersals out of Africa: A Review of the Current State of the Debate.” International Journal of Evolutionary Biology 2011: Article ID 615094. https://doi.org/10.4061/2011/615094.
Brown, Samantha, Thomas Higham, Viviane Slon, Svante Pääbo, Matthias Meyer, Katerina Douka, Fiona Brock, et al. 2016. “Identification of a New Hominin Bone from Denisova Cave, Siberia, Using Collagen Fingerprinting and Mitochondrial DNA Analysis.” Science Reports 6: 23559. https://doi.org/10.1038/srep23559.
Green, Richard E., Johannes Krause, Adrian W. Briggs, Tomislav Maricic, Udo Stenzel, Martin Kircher, Nick Patterson, et al. 2010. “A Draft Sequence of the Neanderthal Genome.” Science 328 (5979): 710–722.
Krings, Matthias, Anne Stone, Ralf W. Schmitz, Heike Krainitzki, Mark Stoneking, and Svante Pääbo. 1997. “Neanderthal DNA Sequences and the Origin of Modern Humans.” Cell 90 (1): 19–30.
Meyer, Matthias, Juan-Luis Arsuaga, Cesare de Filippo, Sarah Nagel, Ayinuer Aximu-Petri, Birgit Nickel, Ignacio Martínez, et al. 2016. “Nuclear DNA Sequences from the Middle Pleistocene Sima de los Huesos Hominins.” Nature 531: 504–507.
Pinhasi, Ron, Daniel Fernandes, Kendra Sirak, Mario Novak, Sarah Connell, Songül Alpaslan-Roodenberg, Fokke Gerritsen, et al. 2015. “Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone.” PLoS One 10 (6): e0129102. https://doi.org/10.1371/journal.pone.0129102.
Prüfer, Kay, Fernando Racimo, Nick Patterson, Flora Jay, Sriram Sankararaman, Susanna Sawyer, Anja Heinze, et al. 2014. “The Complete Genome Sequence of a Neanderthal from the Altai Mountains.” Nature 505 (7481): 43–49.
Prüfer, Kay, Cesare De Filippo, Steffi Grote, Fabrizio Mafessoni, Petra Korlević, Mateja Hajdinjak, Benjamin Vernot, et al. 2017. “A High-Coverage Neandertal Genome from Vindija Cave in Croatia.” Science 358 (6363): 655–658.
Sankararaman, Sriram, Swapan Mallick, Nick Patterson, and David Reich. 2016. “The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans.” Current Biology 26 (9): 1241–1247.
These are genomic sequences which have been sequenced multiple times to ensure that the sequence produced is a true reflection of the genomic sequence, and reduce the likelihood that the sequence has sequencing errors as a result of the sequencing process.
A form of ancient DNA in which DNA originates from the specimen being examined.
DNA that originates from sources outside of the specimen you are trying to sequence.
The chemical process that results in the conversion of Cytosine to uracil, which results in Cytosine to Thymine conversions during sequencing.
A nucleic acid strand that terminates at the chemical group attached to the fifth carbon in the sugar-ring.
A nucleic acid strand that terminates at the hydroxyl (-OH) chemical group attached to the third carbon in the sugar-ring.
DNA sequencing technologies developed in the early 21st century that are capable of sequencing many DNA molecules at a time.
A set of genetic variants located on a single stretch of the genome. This unique combination of variants on a stretch of the genome can be used to differentiate groups that will have different combinations of variants.
These are models which allow for inference of how genetic variants sampled from a population may have originated from a common ancestor.
These are changes that occur in the protein-coding region of the genome and result in a change in amino acid sequence of the protein being produced.
Mutations that occur in the protein-coding region of the genome but do not result in a change in amino acid sequence of the protein being produced.
A measure of how many genes within a diploid genome are made up of more than one variant for a gene.
A nonidentical DNA sequence found in the same gene location on a homologous chromosome, or gene copy, that codes for the same trait but produces a different phenotype.
A genotype comprising an identical set of alleles.
A term often used to describe gene flow between nonhuman populations.
A cellular process that occurs during meiosis I in which homologous chromosomes pair up and sister chromatids on different chromosomes physically swap genetic information.
This is the movement of genes from one species to the gene pool of another species through hybridization between the species and backcross into the parental population by hybrid offspring.
A measure of how long two genomic sequences have been changing independently.
